植物CRISPR/Cas9系統(tǒng)載體基因敲除/基因組DNA編輯技術(shù)
- 價(jià) 格:¥8920
- 貨 號(hào):BioVector-PlantCas9
- 產(chǎn) 地:北京
- BioVector NTCC典型培養(yǎng)物保藏中心
- 聯(lián)系人:Dr.Xu, Biovector NTCC Inc.
電話:400-800-2947 工作QQ:1843439339 (微信同號(hào))
郵件:Biovector@163.com
手機(jī):18901268599
地址:北京
- 已注冊
CRISPR-Cas系統(tǒng)是繼鋅指核酸酶(ZFNs)和TALEN核酸酶之后的另一個(gè)可精確定點(diǎn)編輯基因組DNA的新技術(shù),具有設(shè)計(jì)構(gòu)建簡單快速等優(yōu)點(diǎn)。已在人類細(xì)胞系、斑馬魚、小鼠、果蠅和酵母等多個(gè)物種中利用。最近,國內(nèi)外研究人員紛紛利用CRISPR-Cas系統(tǒng)定點(diǎn)突變了水稻、小麥、擬南芥等多種作物的特定基因,都證實(shí)CRISPR-Cas系統(tǒng)能夠用于植物的基因組編輯。與ZFN 和TALEN 的設(shè)計(jì)復(fù)雜和各種實(shí)驗(yàn)室試驗(yàn)相比,crispr或許相對(duì)簡單點(diǎn)。crispr/Cas系統(tǒng)允許個(gè)性化的小的非編碼RNA來介導(dǎo)靶標(biāo)基因組DNA的切割,從而使基因能夠通過非同源末端結(jié)合和同源接到的修復(fù)機(jī)制來修飾基因。圖1是利用CRISPR系統(tǒng)敲除植物特定基因的流程簡圖。
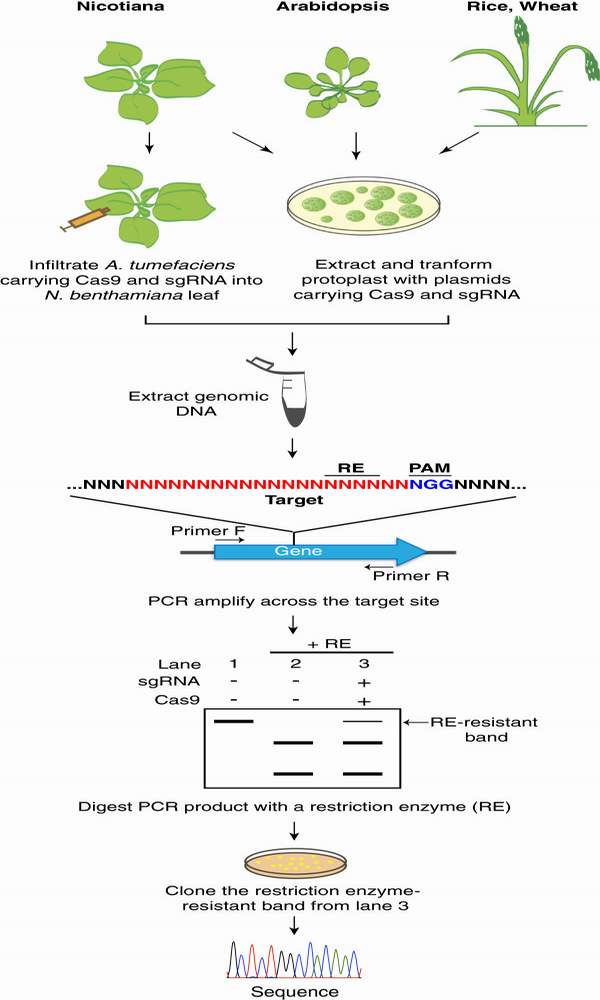
圖1
CRISPR/Cas由20個(gè)堿基的guide RNA 通過Waston-Crick配對(duì)來識(shí)別DNA靶位點(diǎn)序列,從而介導(dǎo)基因的編輯。但是20個(gè)堿基guide RNA也可能與高度同源的DNA序列結(jié)合,從而造成脫靶的問題,這種潛在問題會(huì)限制該技術(shù)在實(shí)際農(nóng)業(yè)生產(chǎn)上的應(yīng)用。但中科院等研究團(tuán)隊(duì)在轉(zhuǎn)基因植株中細(xì)致的分析了靶位點(diǎn)的高度同源位點(diǎn),并未發(fā)現(xiàn)他們跟靶位點(diǎn)一樣發(fā)生突變。同時(shí),經(jīng)過高通量測序,在整個(gè)基因組水平上也未發(fā)現(xiàn)有脫靶問題。據(jù)此,他們認(rèn)為在植物系統(tǒng)中CRISPR/Cas并不像鳥槍或者集束炸彈那樣會(huì)有脫靶問題,而是像巡航導(dǎo)彈一般準(zhǔn)確命中目標(biāo),同時(shí)避免了傷害無辜。
植物CRISPR專家系統(tǒng)服務(wù)于廣大農(nóng)業(yè)科研人員,本系統(tǒng)基于農(nóng)桿菌轉(zhuǎn)化系統(tǒng),含一個(gè)表達(dá)Cas9的二元穿梭載體和一個(gè)表達(dá)sgRNA的中間載體,二元穿梭載體質(zhì)粒架構(gòu)基于植物表達(dá)載體pCambia 1303,sgRNA表達(dá)質(zhì)粒啟動(dòng)子為水稻U3 promoter,Cas9基因?yàn)樗久艽a子優(yōu)化且?guī)蓚€(gè)NLS。
根癌農(nóng)桿菌是一種能誘發(fā)植物產(chǎn)生腫瘤的細(xì)菌,根癌農(nóng)桿菌中誘導(dǎo)植物產(chǎn)生腫瘤的質(zhì)粒(Tumor inducing plasmid),簡稱為 Ti 質(zhì)粒。野生型農(nóng)桿菌的 Ti 質(zhì)粒,含有兩個(gè)與致瘤有關(guān)的區(qū)域:一個(gè)是 T-DNA 區(qū)(transferred DNA region),含致瘤基因;另一個(gè)是毒性區(qū)(Virulence region),在 T-DNA 的切割、轉(zhuǎn)移與整合過程中起作用。用于植物基因轉(zhuǎn)化的農(nóng)桿菌 Ti 質(zhì)粒載體系統(tǒng)的構(gòu)建,是將野生 Ti質(zhì)粒中的致瘤基因刪除,并在 T-DNA 區(qū)域內(nèi)插入適當(dāng)?shù)倪x擇標(biāo)記和多克隆位點(diǎn)。圖2、3是sgRNA表達(dá)質(zhì)粒載體圖;圖4是植物表達(dá)載體 pPL-rCas9的 DNA 圖譜,以 CaMV35S 啟動(dòng)子驅(qū)動(dòng)的水稻密碼子優(yōu)化的Cas9基因以潮霉素抗性為選擇標(biāo)記基因,以β-葡萄糖苷酶(β-glucuronidase,Gus)為報(bào)告基因,經(jīng)此質(zhì)粒轉(zhuǎn)化的獲得的轉(zhuǎn)基因細(xì)胞、組織或植株,具有抗卡那霉素的特性,經(jīng)組織化學(xué)染色呈藍(lán)色。當(dāng)構(gòu)建好sgRNA質(zhì)粒后只要用Kpn I單酶切后一步連接入pPL-rCas9質(zhì)粒即可用于下游的植物CRISPR基因敲除工程操作。
1、構(gòu)建sgRNA表達(dá)載體
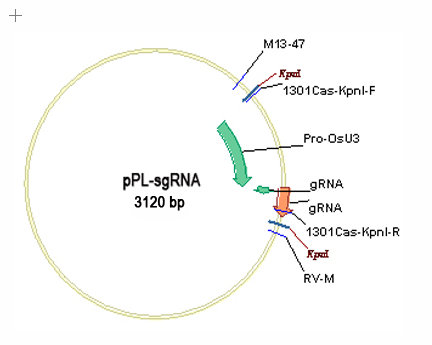
圖2
2、Kpn I單酶切線性化pPL-sgRNA載體
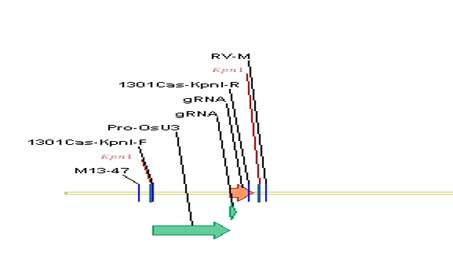
圖3
3、插入到經(jīng)Kpn I 單酶切線性化的pPL-rCas9的載體,得到靶向特定基因的CRISPR基因敲除質(zhì)粒。
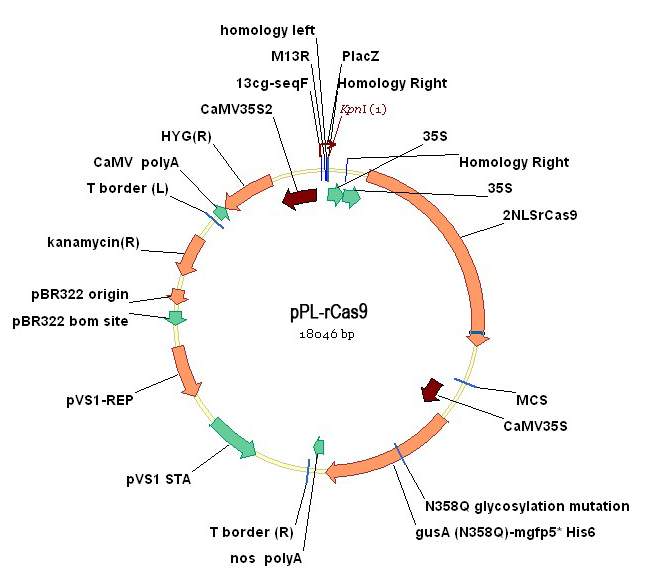
圖4
4、轉(zhuǎn)化農(nóng)桿菌及其下游基因工程篩選操作
參考文獻(xiàn):
1. Belhaj, K., Chaparro-Garcia, A., Kamoun,S., and Nekrasov, V. 2013. Plant genome editing made easy: targeted mutagenesisin model and crop plants using the CRISPR/Cas system. Plant Methods. 9:39.
2. Bos,J. I. B., Kanneganti, T.-D., Young, C., Cakir, C., Huitema, E., Win, J., et al.2006. The C-terminal half of Phytophthora infestans RXLR effector AVR3a issufficient to trigger R3a-mediated hypersensitivity and suppress INF1-inducedcell death in Nicotiana benthamiana. The Plant Journal. 48:165–176.
3. Carroll,D. 2013. Staying on target with CRISPR-Cas. Nat Biotech. 31:807–809.
4. D’Halluin,K., Vanderstraeten, C., Van Hulle, J., Rosolowska, J., Van Den Brande, I.,Pennewaert, A., et al. 2013. Targeted molecular trait stacking in cottonthrough targeted double-strand break induction. Plant Biotechnology Journal.11:933–941.
5. Deltcheva,E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., etal. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factorRNase III. Nature. 471:602–607.
6. Fauser,F., Roth, N., Pacher, M., Ilg, G., Sánchez-Fernández, R., Biesgen, C., et al.2012. In planta gene targeting. PNAS. 109:7535–7540.
7. Feng,Z., Zhang, B., Ding, W., Liu, X., Yang, D.-L., Wei, P., et al. 2013. Efficientgenome editing in plants using a CRISPR/Cas system. Cell Res. 23:1229–1232.
8. Vander Hoorn, R. A. L., Laurent, F., Roth, R., and De Wit, P. J. G. M. 2000.Agroinfiltration Is a Versatile Tool That Facilitates Comparative Analyses of Avr 9/ Cf-9 -Induced and Avr 4/ Cf-4 -Induced Necrosis.Molecular Plant-Microbe Interactions. 13:439–446.
9. Hsu,P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V.,et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. NatBiotech. 31:827–832.
10. Jiang, W., Zhou, H., Bi, H., Fromm, M.,Yang, B., and Weeks, D. P. 2013. Demonstration of CRISPR/Cas9/sgRNA-mediatedtargeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucl.Acids Res. 41:e188–e188.
11. Kuzma, J., and Kokotovich, A. 2011.Renegotiating GM crop regulation. EMBO reports. 12:883–888.
12. Li, J.-F., Norville, J. E., Aach, J.,McCormack, M., Zhang, D., Bush, J., et al. 2013. Multiplex and homologousrecombination-mediated genome editing in Arabidopsis and Nicotiana benthamianausing guide RNA and Cas9. Nat Biotech. 31:688–691.
13. Lusser, M., and Davies, H. V. 2013. Comparativeregulatory approaches for groups of new plant breeding techniques. NewBiotechnology. 30:437–446.
14. Lusser, M., Parisi, C., Plan, D., andRodríguez-Cerezo, E. 2012. Deployment of new biotechnologies in plant breeding.Nat Biotech. 30:231–239.
15. Mali, P., Aach, J., Stranges, P. B.,Esvelt, K. M., Moosburner, M., Kosuri, S., et al. 2013. CAS9 transcriptionalactivators for target specificity screening and paired nickases for cooperativegenome engineering. Nat Biotech. 31:833–838.
16. Mao, Y., Zhang, H., Xu, N., Zhang, B.,Gou, F., and Zhu, J.-K. 2013. Application of the CRISPR-Cas System forEfficient Genome Engineering in Plants. Molecular Plant. 6:2008–2011.
17. Miao, J., Guo, D., Zhang, J., Huang, Q.,Qin, G., Zhang, X., et al. 2013. Targeted mutagenesis in rice using CRISPR-Cassystem. Cell Res. 23:1233–1236.
18. Mussolino, C., and Cathomen, T. 2013. RNAguides genome engineering. Nat Biotech. 31:208–209.
19. Nekrasov, V., Staskawicz, B., Weigel, D.,Jones, J. D. G., and Kamoun, S. 2013. Targeted mutagenesis in the model plantNicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotech. 31:691–693.
20. Pattanayak, V., Lin, S., Guilinger, J.P., Ma, E., Doudna, J. A., and Liu, D. R. 2013. High-throughput profiling ofoff-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. NatBiotech. 31:839–843.
21. Qi, Y., Zhang, Y., Zhang, F., Baller, J.A., Cleland, S. C., Ryu, Y., et al. 2013. Increasing frequencies ofsite-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNArepair pathways. Genome Res. 23:547–554.
22. Shan, Q., Wang, Y., Li, J., Zhang, Y.,Chen, K., Liang, Z., et al. 2013. Targeted genome modification of crop plantsusing a CRISPR-Cas system. Nat Biotech. 31:686–688.
23. Voytas, D. F. 2013. Plant GenomeEngineering with Sequence-Specific Nucleases. Annual Review of Plant Biology.64:327–350.
24. Weber, E., Engler, C., Gruetzner, R.,Werner, S., and Marillonnet, S. 2011. A Modular Cloning System for StandardizedAssembly of Multigene Constructs. PLoS ONE. 6:e16765.
CRISPR/Cas9相關(guān)產(chǎn)品:
Precut pSG-target Cloning kit & SSA assay
Different type cas9 expression plasmid CRISPR /Cas9載體
T7 sgRNA MICscriptTM KIT/sgRNA synthesis product
Precut SgRNA Cloning kit & pSD-gRNA Plasmid
EzOmicsTM RNA QUICK clear KIT-------一步純化RNA轉(zhuǎn)錄產(chǎn)物
CRISPR相關(guān)服務(wù):
sgRNA序列設(shè)計(jì)
sgRNA表達(dá)載體構(gòu)建
sgRNA體外合成
全基因組sgRNA文庫的構(gòu)建
sgRNA活性檢測
Surveyor法及測序驗(yàn)證
穩(wěn)定表達(dá)Cas9蛋白細(xì)胞株篩選
利用CRISPR系統(tǒng)進(jìn)行基因重組donor質(zhì)粒構(gòu)建
植物CRISPR
您正在向 biovector.net 發(fā)送關(guān)于產(chǎn)品 植物CRISPR/Cas9系統(tǒng)載體基因敲除/基因組DNA編輯技術(shù) 的詢問
- 公告/新聞